| Applications: | ELISA |
| Reactivity: | Mouse |
| Note: | FOR SCIENTIFIC EDUCATIONAL RESEARCH USE ONLY (RUO). MUST NOT BE USED IN DIAGNOSTIC OR OTHER MEDICAL APPLICATIONS. |
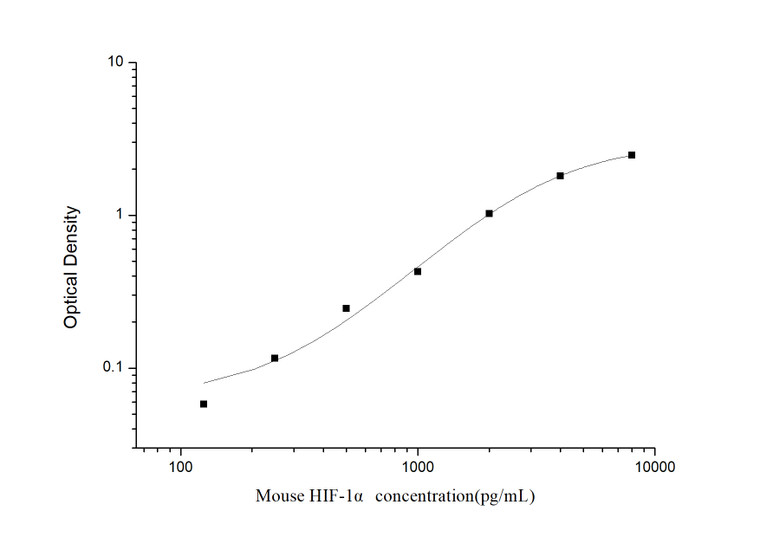
| Sensitivity: | 75pg/mL |
| Detection Limit: | 125~8000pg/mL |
| Short Description: | This Hif1a Sandwich ELISA is an in-vitro enzyme-linked immunosorbent assay for the measurement of samples in mouse cell culture supernatant, serum and plasma (EDTA, citrate, heparin). |
| Storage Instruction: | If unopened the kit may be stored at 2-8°C for up to 1 month. If the kit will not be used within 1 month, store the components separately, according to the component table in the manual. |
| Assay Time: | 3.5h |
| Detection: | Colormetric |
| Gene Symbol: | Hif1a |
| Gene ID: | 15251 |
| Uniprot ID: | HIF1A_MOUSE |
| Specificity: | This kit recognizes Mouse HIF-1 Alpha in samples. No significant cross-reactivity or interference between Mouse HIF-1 Alpha and analogues was observed. |
| Sample Type: | Serum, plasma and other biological fluids |
| Post Translational Modifications | S-nitrosylation of Cys-810 may be responsible for increased recruitment of p300 coactivator necessary for transcriptional activity of HIF-1 complex. Requires phosphorylation for DNA-binding. Phosphorylation at Ser-247 by CSNK1D/CK1 represses kinase activity and impairs ARNT binding. Phosphorylation by GSK3-beta and PLK3 promote degradation by the proteasome. Sumoylated.with SUMO1 under hypoxia. Sumoylation is enhanced through interaction with RWDD3. Both sumoylation and desumoylation seem to be involved in the regulation of its stability during hypoxia. Sumoylation can promote either its stabilization or its VHL-dependent degradation by promoting hydroxyproline-independent HIF1A-VHL complex binding, thus leading to HIF1A ubiquitination and proteasomal degradation. Desumoylation by SENP1 increases its stability amd transcriptional activity. There is a disaccord between various publications on the effect of sumoylation and desumoylation on its stability and transcriptional activity (Probable). Acetylation of Lys-545 by ARD1 increases interaction with VHL and stimulates subsequent proteasomal degradation. Deacetylation of Lys-719 by SIRT2 increases its interaction with and hydroxylation by EGLN1 thereby inactivating HIF1A activity by inducing its proteasomal degradation. Ubiquitinated.in normoxia, following hydroxylation and interaction with VHL. Lys-545 appears to be the principal site of ubiquitination. Clioquinol, the Cu/Zn-chelator, inhibits ubiquitination through preventing hydroxylation at Asn-813. Ubiquitinated by E3 ligase VHL. Deubiquitinated by UCHL1. The iron and 2-oxoglutarate dependent 3-hydroxylation of asparagine is (S) stereospecific within HIF CTAD domains. In normoxia, is hydroxylated on Pro-402 and Pro-577 in the oxygen-dependent degradation domain (ODD) by EGLN1/PHD2 and EGLN2/PHD1. EGLN3/PHD3 has also been shown to hydroxylate Pro-577. The hydroxylated prolines promote interaction with VHL, initiating rapid ubiquitination and subsequent proteasomal degradation. Deubiquitinated by USP20. Under hypoxia, proline hydroxylation is impaired and ubiquitination is attenuated, resulting in stabilization. In normoxia, is hydroxylated on Asn-813 by HIF1AN, thus abrogating interaction with CREBBP and EP300 and preventing transcriptional activation. Repressed by iron ion, via Fe(2+) prolyl hydroxylase (PHD) enzymes-mediated hydroxylation and subsequent proteasomal degradation. |
| Function | Functions as a master transcriptional regulator of the adaptive response to hypoxia. Under hypoxic conditions, activates the transcription of over 40 genes, including erythropoietin, glucose transporters, glycolytic enzymes, vascular endothelial growth factor, HILPDA, and other genes whose protein products increase oxygen delivery or facilitate metabolic adaptation to hypoxia. Plays an essential role in embryonic vascularization, tumor angiogenesis and pathophysiology of ischemic disease. Heterodimerizes with ARNT.heterodimer binds to core DNA sequence 5'-TACGTG-3' within the hypoxia response element (HRE) of target gene promoters. Activation requires recruitment of transcriptional coactivators such as CREBBP and EP300. Activity is enhanced by interaction with NCOA1 and/or NCOA2. Interaction with redox regulatory protein APEX1 seems to activate CTAD and potentiates activation by NCOA1 and CREBBP. Involved in the axonal distribution and transport of mitochondria in neurons during hypoxia. |
| Protein Name | Hypoxia-Inducible Factor 1-Alpha Hif-1-Alpha Hif1-Alpha Arnt-Interacting Protein |
| Database Links | Reactome: R-MMU-1234158 Reactome: -MMU-1234174 Reactome: -MMU-1234176 Reactome: -MMU-5689880 Reactome: -MMU-8857538 Reactome: -MMU-8951664 |
| Cellular Localisation | Cytoplasm Nucleus Nucleus Speckle Colocalizes With Hif3a Isoform 2 In The Nucleus And Speckles Cytoplasmic In Normoxia Nuclear Translocation In Response To Hypoxia |
| Alternative ELISA Names | Hypoxia-Inducible Factor 1-Alpha ELISA kit Hif-1-Alpha ELISA kit Hif1-Alpha ELISA kit Arnt-Interacting Protein ELISA kit Hif1a ELISA kit |
| Specificity | This kit recognizes Mouse HIF-1α in samples. No significant cross-reactivity or interference between Mouse HIF-1α and analogues was observed. |
| Reproducibility | Both intra-CV and inter-CV are |
Information sourced from Uniprot.org
| Item | Specifications | Storage |
|---|---|---|
| Micro ELISA Plate (Dismountable) | 96T: 8 wells ×12 strips strips | -20℃, 6 months |
| Reference Standard | 96T: 2 vials 48T: 1 vial | -20℃, 6 months |
| Concentrated Biotinylated Detection Ab (100×) | 96T: 1 vial, 120 μL 60 μL | -20℃, 6 months |
| Concentrated HRP Conjugate (100×) | 96T: 1 vial, 120 μL 60 μL | -20℃ (Protect from light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 2-8°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | 2-8°C, 6 months |
| HRP Conjugate Diluent | 1 vial, 14 mL | 2-8°C, 6 months |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | 2-8°C, 6 months |
| Substrate Reagent | 1 vial, 10 mL | 2-8℃ (Protect from light) |
| Stop Solution | 1 vial, 10 mL | 2-8°C |
| Plate Sealer | 5 pieces | |
| Manual | 1 copy | |
| Certificate of Analysis | 1 copy |
| Sample Type | Range (%) | Average Recovery (%) |
|---|---|---|
| Serum(n=8) | 86-100 | 93 |
| EDTA plasma (n=8) | 93-105 | 99 |
| Cell culture media (n=8) | 89-102 | 95 |
| Intra-assay Precision | Intra-assay Precision | Intra-assay Precision | Inter-assay Precision | Inter-assay Precision | Inter-assay Precision | |
|---|---|---|---|---|---|---|
| Sample | 1.00 | 2.00 | 3.00 | 1.00 | 2.00 | 3.00 |
| n | 20.00 | 20.00 | 20.00 | 20.00 | 20.00 | 20.00 |
| Mean (pg/mL) | 387.70 | 1086.20 | 2742.50 | 371.80 | 1120.30 | 2531.80 |
| Standard deviation | 24.80 | 46.70 | 109.70 | 19.70 | 58.30 | 108.90 |
| CV (%) | 6.40 | 4.30 | 4.00 | 5.30 | 5.20 | 4.30 |