| Host: |
HEK293 cells |
| Reactivity: |
Human |
| Note: |
STRICTLY FOR FURTHER SCIENTIFIC RESEARCH USE ONLY (RUO). MUST NOT TO BE USED IN DIAGNOSTIC OR THERAPEUTIC APPLICATIONS. |
| Short Description: |
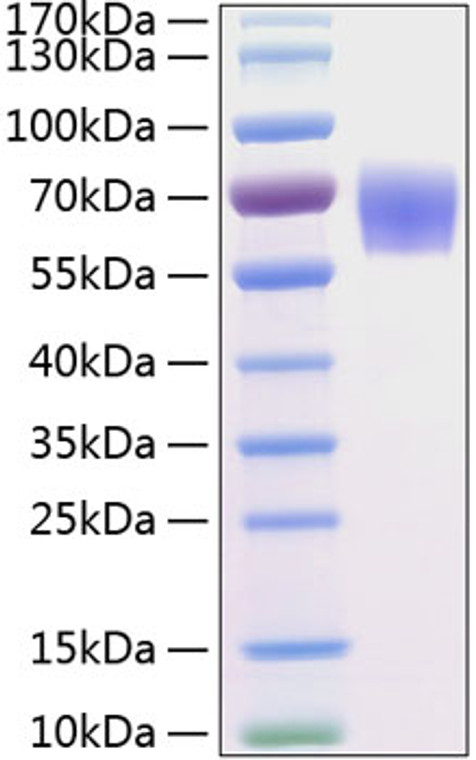
Recombinant-Human BACE-1/ASP2-C-His protein was developed from hek293 cells and has a target region of C-His. For use in research applications. |
| Formulation: |
Lyophilized from a 0.22 Mu m filtered solution of PBS, pH 7.4. Contact us for customized product form or formulation. |
| Immunoreactivity: |
Measured by its ability to cleave a fluorogenic peptide substrate, Mca-SEVNLDAEFRK (Dpn) RR-NH2. The specific activity is >5.5 pmol/min/Mu g. |
| Gene Symbol: |
BACE1 |
| Gene ID: |
23621 |
| Uniprot ID: |
BACE1_HUMAN |
| Immunogen: |
Recombinant Human BACE-1/ASP2 Protein is produced by HEK293 expression system. The target protein is expressed with sequence (Thr22-Tyr460) of human BACE1/ASP2 (Accession #NP_036236.1) fused with an 8×His tag at the C-terminus. |
| Tissue Specificity | Expressed at high levels in the brain and pancreas. In the brain, expression is highest in the substantia nigra, locus coruleus and medulla oblongata. |
| Post Translational Modifications | N-Glycosylated. Addition of a bisecting N-acetylglucosamine by MGAT3 blocks lysosomal targeting, further degradation and is required for maintaining stability under stress conditions. Acetylated in the endoplasmic reticulum at Lys-126, Lys-275, Lys-279, Lys-285, Lys-299, Lys-300 and Lys-307. Acetylation by NAT8 and NAT8B is transient and deacetylation probably occurs in the Golgi. Acetylation regulates the maturation, the transport to the plasma membrane, the stability and the expression of the protein. Palmitoylation mediates lipid raft localization. Ubiquitinated at Lys-501, ubiquitination leads to lysosomal degradation. Monoubiquitinated and 'Lys-63'-linked polyubitinated. Deubiquitnated by USP8.inhibits lysosomal degradation. Phosphorylation at Ser-498 is required for interaction with GGA1 and retrograded transport from endosomal compartments to the trans-Golgi network. Non-phosphorylated BACE1 enters a direct recycling route to the cell surface. |
| Function | Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase. Cleaves CHL1. |
| Protein Name | Beta-Secretase 1Aspartyl Protease 2Asp2Asp 2Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1Beta-Site App Cleaving Enzyme 1Memapsin-2Membrane-Associated Aspartic Protease 2 |
| Database Links | Reactome: R-HSA-977225 |
| Cellular Localisation | Cell MembraneSingle-Pass Type I Membrane ProteinGolgi ApparatusTrans-Golgi NetworkEndoplasmic ReticulumEndosomeCell SurfaceCytoplasmic Vesicle MembraneMembrane RaftLysosomeLate EndosomeEarly EndosomeRecycling EndosomeCell ProjectionAxonDendritePredominantly Localized To The Later Golgi/Trans-Golgi Network (Tgn) And Minimally Detectable In The Early Golgi CompartmentsA Small Portion Is Also Found In The Endoplasmic ReticulumEndosomes And On The Cell SurfaceColocalization With App In Early Endosomes Is Due To Addition Of Bisecting N-Acetylglucosamine Wich Blocks Targeting To Late Endosomes And LysosomesRetrogradly Transported From Endosomal Compartments To The Trans-Golgi Network In A Phosphorylation- And Gga1- Dependent Manner |
| Alternative Protein Names | Beta-Secretase 1 proteinAspartyl Protease 2 proteinAsp2 proteinAsp 2 proteinBeta-Site Amyloid Precursor Protein Cleaving Enzyme 1 proteinBeta-Site App Cleaving Enzyme 1 proteinMemapsin-2 proteinMembrane-Associated Aspartic Protease 2 proteinBACE1 proteinBACE proteinKIAA1149 protein |
Information sourced from Uniprot.org
12 months for antibodies. 6 months for ELISA Kits. Please see website T&Cs for further guidance