| Applications: |
ELISA |
| Reactivity: |
Human |
| Note: |
STRICTLY FOR FURTHER SCIENTIFIC RESEARCH USE ONLY (RUO). MUST NOT TO BE USED IN DIAGNOSTIC OR THERAPEUTIC APPLICATIONS. |
| Sensitivity: |
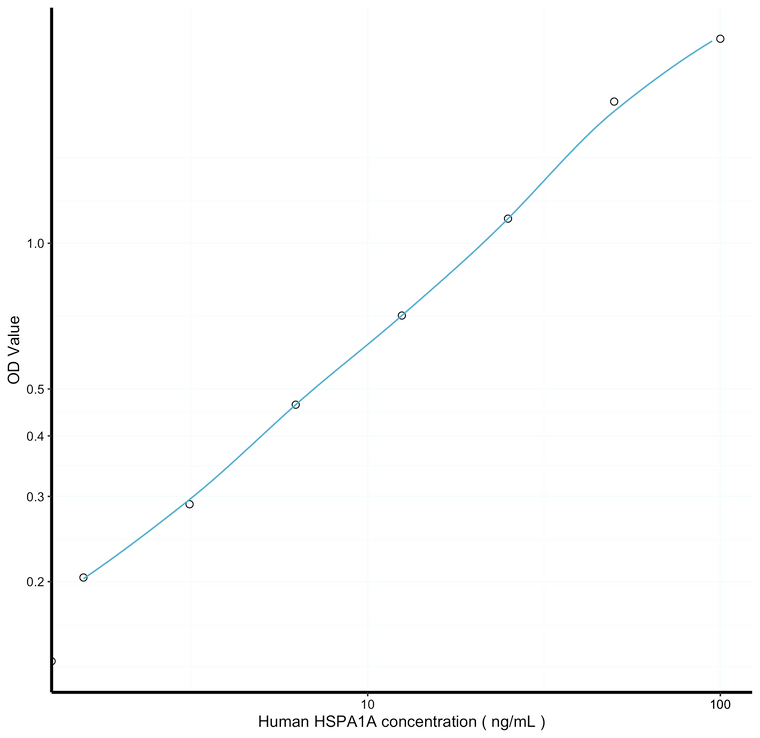
0.54ng/mL |
| Detection Limit: |
1.56-100ng/mL |
| Short Description: |
This HSPA1A Sandwich ELISA Kit, Ready-To-Use is an in-vitro enzyme-linked immunosorbent assay for the measurement of samples in human cell culture supernatant, serum and plasma (EDTA, citrate, heparin). |
| Storage Instruction: |
The whole kit may be stored at-20°C for up to 12 months from receipt. An unopened kit may be stored in the fridge at 2-8°C for up to 6 months. Once opened store individual kit contents according to components table provided with the kit. |
| Assay Time: |
3 hrs |
| Gene Symbol: |
HSPA1A |
| Gene ID: |
3303/3304 |
| Uniprot ID: |
HS71A_HUMAN |
| Immunogen Region: |
Ready-To-Use |
| Sample Type: |
serum, plasma, tissue homogenates, cell lysates, cell culture supernates or other biological fluids. |
| Tissue Specificity | |
| Post Translational Modifications | In response to cellular stress, acetylated at Lys-77 by NA110 and then gradually deacetylated by HDAC4 at later stages. Acetylation enhances its chaperone activity and also determines whether it will function as a chaperone for protein refolding or degradation by controlling its binding to co-chaperones HOPX and STUB1. The acetylated form and the non-acetylated form bind to HOPX and STUB1 respectively. Acetylation also protects cells against various types of cellular stress. |
| Function | Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, folding and transport of newly synthesized polypeptides, activation of proteolysis of misfolded proteins and the formation and dissociation of protein complexes. Plays a pivotal role in the protein quality control system, ensuring the correct folding of proteins, the re-folding of misfolded proteins and controlling the targeting of proteins for subsequent degradation. This is achieved through cycles of ATP binding, ATP hydrolysis and ADP release, mediated by co-chaperones. The co-chaperones have been shown to not only regulate different steps of the ATPase cycle, but they also have an individual specificity such that one co-chaperone may promote folding of a substrate while another may promote degradation. The affinity for polypeptides is regulated by its nucleotide bound state. In the ATP-bound form, it has a low affinity for substrate proteins. However, upon hydrolysis of the ATP to ADP, it undergoes a conformational change that increases its affinity for substrate proteins. It goes through repeated cycles of ATP hydrolysis and nucleotide exchange, which permits cycles of substrate binding and release. The co-chaperones are of three types: J-domain co-chaperones such as HSP40s (stimulate ATPase hydrolysis by HSP70), the nucleotide exchange factors (NEF) such as BAG1/2/3 (facilitate conversion of HSP70 from the ADP-bound to the ATP-bound state thereby promoting substrate release), and the TPR domain chaperones such as HOPX and STUB1. Maintains protein homeostasis during cellular stress through two opposing mechanisms: protein refolding and degradation. Its acetylation/deacetylation state determines whether it functions in protein refolding or protein degradation by controlling the competitive binding of co-chaperones HOPX and STUB1. During the early stress response, the acetylated form binds to HOPX which assists in chaperone-mediated protein refolding, thereafter, it is deacetylated and binds to ubiquitin ligase STUB1 that promotes ubiquitin-mediated protein degradation. Regulates centrosome integrity during mitosis, and is required for the maintenance of a functional mitotic centrosome that supports the assembly of a bipolar mitotic spindle. Enhances STUB1-mediated SMAD3 ubiquitination and degradation and facilitates STUB1-mediated inhibition of TGF-beta signaling. Essential for STUB1-mediated ubiquitination and degradation of FOXP3 in regulatory T-cells (Treg) during inflammation. Negatively regulates heat shock-induced HSF1 transcriptional activity during the attenuation and recovery phase period of the heat shock response. Involved in the clearance of misfolded PRDM1/Blimp-1 proteins. Sequesters them in the cytoplasm and promotes their association with SYNV1/HRD1, leading to proteasomal degradation. (Microbial infection) In case of rotavirus A infection, serves as a post-attachment receptor for the virus to facilitate entry into the cell. |
| Protein Name | Heat Shock 70 Kda Protein 1aHeat Shock 70 Kda Protein 1Hsp70-1Hsp70.1 |
| Database Links | Reactome: R-HSA-168330Reactome: R-HSA-3371453Reactome: R-HSA-3371497Reactome: R-HSA-3371568Reactome: R-HSA-3371571Reactome: R-HSA-450408Reactome: R-HSA-6798695 |
| Cellular Localisation | CytoplasmNucleusCytoskeletonMicrotubule Organizing CenterCentrosomeSecretedLocalized In Cytoplasmic Mrnp Granules Containing Untranslated Mrnas |
| Alternative ELISA Names | Heat Shock 70 Kda Protein 1a ELISA kitHeat Shock 70 Kda Protein 1 ELISA kitHsp70-1 ELISA kitHsp70.1 ELISA kitHSPA1A ELISA kitHSP72 ELISA kitHSPA1 ELISA kitHSX70 ELISA kit |
| output | |
Information sourced from Uniprot.org
12 months for antibodies. 6 months for ELISA Kits. Please see website T&Cs for further guidance